
Fast Interaction REfinement in molecular DOCKing
About FireDock
The FireDock (ref. 1) server addresses the refinement problem of protein-protein docking solutions. The method simultaneously targets the problem of flexibility and scoring of solutions produced by fast rigid-body docking algorithms. Given a set of up to 1000 potential docking candidates, FireDock refines and scores them according to an energy function, spending about 3.5 seconds per candidate solution. To the best of our knowledge, this is the first webserver that allows performing large-scale flexible refinement and scoring of docking solutions online.
There are two input options for FireDock. In the first option, the user uploads/specifies PDB codes of two PDB files (receptor and ligand) and provides a list of transformations. Each transformation, when applied on the ligand, produces a candidate docking solution. In the second option, the user can upload an input PDB file, with each docking solution represented by a MODEL. The candidate solutions for FireDock can be generated by rigid-body docking methods, such as PatchDock (ref. 2,3), FFT-based methods such as ZDOCK , GRAMM-X , Hex, ClusPro etc. In addition, we provide an option for automatic redirection of solutions from PatchDock webserver.
Each candidate is subsequently refined by restricted interface side-chain rearrangement and by soft rigid-body optimization. The side-chain flexibility is modeled by rotamers and the obtained combinatorial optimization problem is solved by integer linear programming (ref. 4). Following rearrangement of the side-chains, the relative position of the docking partners is refined by Monte Carlo minimization of the binding score function. The refined candidates are ranked by the binding score. This score includes Atomic Contact Energy (ref. 5), softened van der Waals interactions, partial electrostatics and additional estimations of the binding free energy.
The output is a ranked list of all the input solutions. The refined complex structure is generated for up to 100 low-energy candidates. The user can view the complexes in the Jmol applet window and/or download the structures.
The method (and server) has been extensively and successfully tested on protein-protein docking benchmark of ~80 complexes and on all the CAPRI targets.
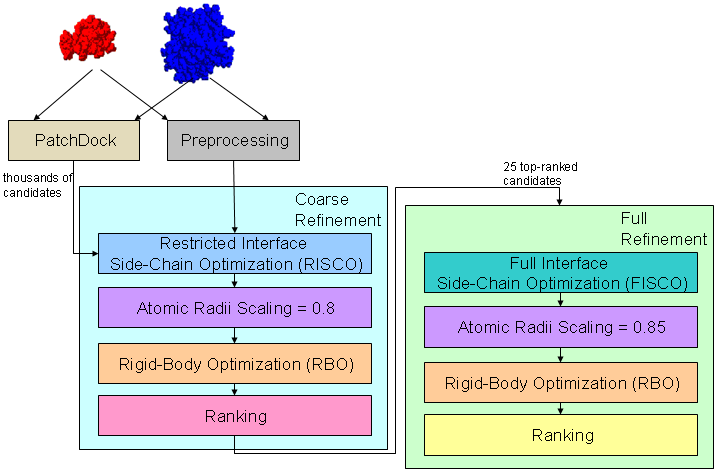
The figure below shows an example of FireDock usage. Transformations of docking candidates are generated by PatchDock (ref. 2,3) and are given as an input to FireDock. First a coarse refinement is performed, using a restricted interface side-chain optimization with atomic radii scaling of 0.8, in order to allow a certain amount of steric clashes. The refined candidates are scored and ranked according to the energy function and are returned as an output. Then, FireDock is run again on the best 25 solutions for a final refinement. In this second run, a full interface side-chain optimization is performed with atomic radii scaling of 0.85, in order to reduce the amount of clashes.
References:
- N. Andrusier, R. Nussinov and H. J. Wolfson. FireDock: Fast Interaction Refinement in Molecular Docking. Proteins 2007, 69(1):139-59.
- D. Duhovny, R. Nussinov, and H. J. Wolfson. Efficient unbound docking of rigid molecules. In R. Guigo and D. Gusfield, editors, Workshop on Algorithms in Bioinformatics, volume 2452, pages 185-200. Springer Verlag, 2002.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov, H. J. Wolfson. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl. Acids. Res. 33: W363-367, 2005.
- C. L. Kingsford, B. Chazelle, and M. Singh. Solving and analyzing side-chain positioning problems using linear and integer programming. Bioinformatics, 21(7):1028-1036, 2005.
- C. Zhang, G. Vasmatzis, J. L. Cornette, and C. DeLisi. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol, 267(3):707-726, April 1997.