
 |
| An Algorithm For Protein Hinge Prediction Using Elastic Network Models |
| [About HingeProt] | [Web Server] | [Help] |
|
Motivation: Proteins are highly flexible molecules. It is common to classify protein motions into shear and hinge motion [1]. Shear motions are very limited and involve large number of residues. On the other hand, hinge motions are similar to rotations around an articulated joint and therefore can be very large. Hinge motion is characterized by large changes in main-chain torsional angles occurring at a localized region, which is called a hinge. Hinge motions usually involve a small number of residues, since even one bond can provide the required rotational freedom. This kind of protein motion is free of packing constraints. When a chain exhibits hinge motion at the region connecting two structural domains, each domain behaves as a rigid body and packing interactions can appear/disappear between the interfaces of those rigid bodies. Hinge motions usually occur upon binding to another molecule, or upon activation/deactivation of the protein. One of the most interesting examples is calmodulin. Upon binding to its ligands, there is large-scale movement of calmodulin involving splitting of one long helix. The total rotation of one domain relative to the other is upwards of 150 degrees (see images below and try the server for PDB codes 4cln and 2bbm, chainA). 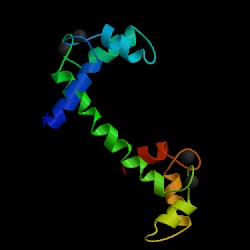 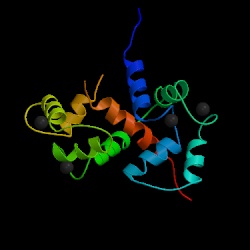
HingeProt [2] is a web server for predicting rigid protein parts and the flexible hinge regions connecting them in the native topology of protein chains by employing elastic network (EN) models. HingeProt makes use of both Gaussian Network Model (GNM) [3, 4] and Anisotropic Network models (ANM) [5]. HingeProt server focuses on the prediction of the rigid parts and the hinge regions using a single static conformation of a protein structure. The hinge regions are the mechanistically informative regions of the structure and are of importance in mediating cooperative motions that have functional importance. GNM calculates the mean-square fluctuations and the correlation between the fluctuations of residues in the most dominant (slowest two) modes, which were shown to overlap with known protein motions. These suggest hinge regions and the cooperation between them. ANM provides the direction of the fluctuations in the corresponding modes. HingeProt is expected to be useful in a range of potential applications, especially in prediction protein-protein association bu flexible docking and in refinement of the structure of the modeled complexes. HingeProt predictions are also helpful in fitting flexible hinge-bent protein structures into EM density maps and refining the EM structures. In addition, hinge regions can help in understanding functional mechanisms of macromolecular structures and assemblies. Given an input protein chain, HingeProt identifies the rigid parts and the hinges connecting them, and the direction of the fluctuation of each residue in the slowest two modes. Method: GNM [3, 4] and its extension ANM [5] are coarse-grained residue level elastic network models. GNM predicts the relative magnitudes of the fluctuations, whereas ANM predicts the directionalities of the collective motions in addition to their magnitudes. GNM results are more robust, and thus are preferentially used for evaluating square displacements in low frequency modes [5]. Here GNM is used to calculate mean-square fluctuations and correlation between the fluctuations of residues, and ANM to generate the conformations that describe the fluctuations of residues from the average, X-ray, structure in the principal directions of motion. In HingeProt, first GNM decomposes the fluctuations of N residues of a structure into a series of N-1 nonzero modes, given the Cartesian coordinates of Ca atoms. The eigenvectors corresponding to the slowest first and second modes are extracted. The square of these vectors describes the mean-square fluctuations (the autocorrelations) of residues from equilibrium positions along the principal coordinates (first and second modes here). Minima of mean square fluctuations at a given mode describe the flexible joints of the structure, i.e. the hinge regions, which connect the rigid units and mobile loops. GNM fluctuations being isotropic by definition, the directions of fluctuations are characterized by ANM. ANM predicts the fluctuations of N residues in the x, y and z directions from the average structure (X-ray or NMR) in 3N-6 ANM nonzero modes. After mapping the ANM modes to GNM modes by comparing the square fluctuations, between the resulting modes in the two models, the directions of the fluctuations of residues in the slowest first and second modes of GNM are obtained by ANM analysis. As the fluctuations are symmetric with respect to the equilibrium positions, ANM predicted deformed structures could be obtained by adding and subtracting the fluctuations of each residue to/from its equilibrium position. For more detailed background of the work see the article.
References : |
For comments: ppdock@tau.ac.il