 |
|
| Structural and Physico-Chemical
Alignment of Protein-Protein Interfaces |
[About I2I-SiteEngine][Server Help] |
| Interface-to-Interface
(I2I)-SiteEngine compares pairs of interacting protein binding sites. |
Below is a detailed description of the main forms used in the
SiteEngine server.
To speed up the search please click on the stage on interest.
Forms and stages of the I2I-SiteEngine
server:
Stage1 - Input complexes definition
Stage2 - Selection of the interacting chains that determine the interface of interest
Stage3 - Process of I2I-SiteEngine
Stage4 - Output of I2I-SiteEngine
Stage1 - Input
molecules definition:
The first stage in activating the I2I-SiteEngine is the definition of the
complexes of interest. The definition is performed through the form
below.
If the molecules are
available in the Protein Data Bank (PDB) the PDB codes are to be
specified, otherwise the molecules of interest can be uploaded to our
server. Using the PDB codes speeds up the process, since no file
transfer is required.

Stage2 - Interface definition
The method automatically extracts all pairs of interacting protein chains and prompts the user to select the chains of interest. Each pair of the presented interacting protein chains (separated by a semicolon) define a potential protein-protein interface. I2I-SiteEngine will compare only the interfaces defined by the selected chains.
Single chain protein molecules that have no protein binding partner in the submitted PDB file can not be compared by the I2I-SiteEngine method

Stage3 - Process of I2I-SiteEngine:
This window shows the process of activation of SiteEngine. The
five main stages are presented and those that are complete are checked
in the checkbox.
In most of the cases the most time consuming stages are the
construction of the surfaces.

Stage4 - Output of I2I-SiteEngine:
This window presents the output of the SiteEngine algorithm.
The correspondence between the chains of the two interfaces is
a-priori unknown, e.g. given two interfaces determined by chains Z:I
and E:I respectively, we have to determined whether the binding site
of chain Z in one interface is more similar to that of chain E or
chain I in the other. I2I-SiteEngine checks both options and selects
the highest ranking correspondence. The output of I2I-SiteEngine is
the list of the matching functional groups of the corresponding
binding sites that constitute the interfaces. Suppose that in the
example above we have recognized that the correct correspondence is to
align the binding site chain Z in one interface to chain E in the
other. Then the output of I2I-SiteEngine will first present the
"Match List of Side 1", i.e. the functional groups common to the
aligned binding sites of chains E and Z. The it will present the
"Match List of Side 2", which will contains the matching functional
groups recognized in the binding site of chains I in both interfaces.
The 10 top ranking solutions are presented. These represent the
highest scoring ways of superimposition of the interfaces of
interest. The file aligned.pdb is the superimposition of the two
complexes by the transformation recognized by I2I-SiteEngine. It can be
either downloaded of viewed directly from the browser. The file
contains the superimposition of the two complexes as well as the
functional groups that are recognized to be shared by the
regions. These are also detailed in the output table. To view these
features in the rasmol view please use the following rasmol script.
The output table of
SiteEngine presents the details of the common functional groups.
Below is the description of the columns of the table (click on the
column of interest to jump to a description):
Chain.ID
AminoAcid
Property
Source
Dist
Conserved AA
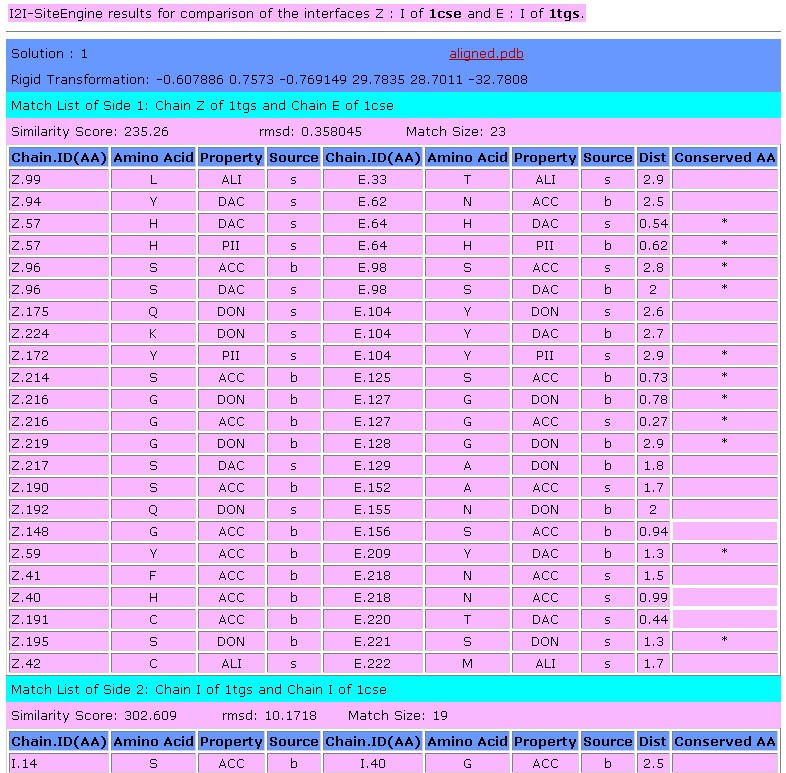
Chain.ID:
The protein chain, followed by the
identity of the amino acid
AminoAcid:
The one
letter amino acid code. However it must be noted that the method is
based on the physico-chemical properties and does not consider the
identity of the amino acids. These are only displayed for the
convenience of analysis.
Property:
The
physico-chemical property that is matched by the algorithm. The method
is based on a representation of each amino acid of a protein as a set
of features that are important for its interaction with other
molecules. The abbreviations of these features are:
DON - Hydrogen bond donor
ACC - Hydrogen bond acceptor
DAC - Hydrogen bond donor and
acceptor (e.g in histidine)
ALI - Aliphatic Hydrophobic property
PII - Aromatic property (pi
contacts)
Source:
This field specifies whether the matched
property is contributed by the backbone or the side-chain of the amino
acid.
The abbreviations are:
b - feature contributed by the backbone
s - feature contributed by the
backbone
Dist:
The distance in space measured
between the matched features.
Conserved
AA:
Marks the features shared by the two molecules that are
contributed by residues with the same identity of the amino acid.
Contact: shulmana@tau.ac.il
Reference:
Shulman-Peleg
A, Mintz S,Nussinov R, Wolfson HJ, J Mol Biol. 2004 Jun
4;339(3):607-33. Protein-Protein
Interfaces:
Recognition of Similar Spatial and Chemical Organizations, Accepted
to WABI-4th
Workshop on Algorithms in
Bioinformatics, LNCS,
Norway, Sep. 14-17 [PDF].





