[About MAPPIS]
[Server Help]
[Download Software]
| |
|
 |
[Home] [About MAPPIS] [Server Help] [Download Software] |
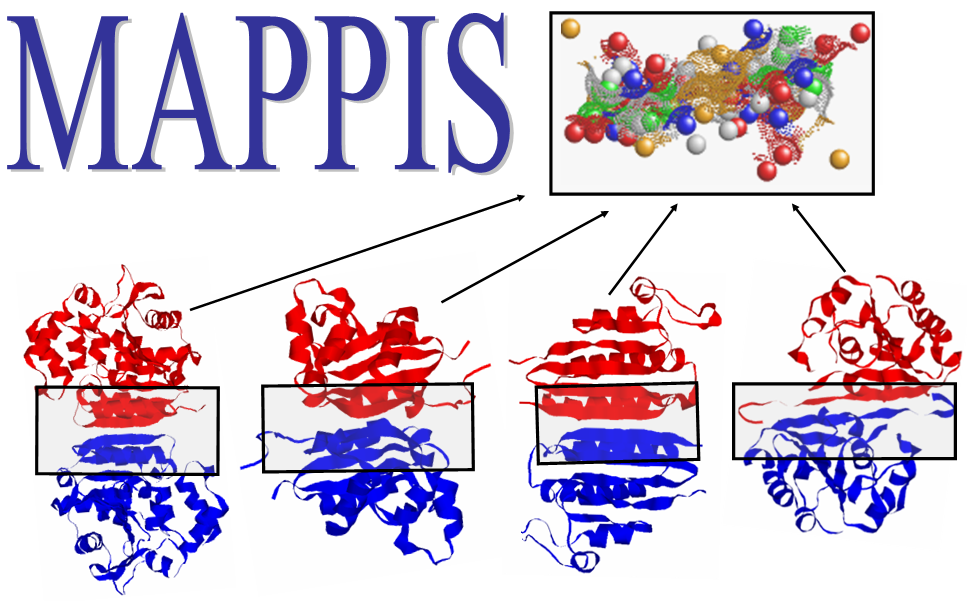
| Multiple Alignment of Protein-Protein InterfaceS (PPIs) | |
| Recognizes spatially conserved chemical interactions shared by a set of PPIs | |

1 - If the chain IDs are known use the following format [PDB] [chain 1]:[chain 2]:
e.g. 1cse E:I, 1cbw HG:I
This will compare between two interfaces:
PPI 1 - created by chains E and I of 1cse
PPI 2 - created by chains HG interacting with chain I in 1cbw
2 - If the interacting chains are unknown type the PDB codes separated by spaces.
e.g.1cse, 1tgs, 1acb
The advantage of the first option is that it allows to define complex interfaces created by more than a pair of proteins chains (for example chains HG of 1cbw and chain I of 1cbw). However, you should not use it if you are not sure that the chains you type actually interact with each other.
If the second option is used and no chains are specified by the user the method automatically extracts all pairs of interacting protein chains and prompts the user to select the chains of interest.

Each pair of the presented interacting protein chains (separated by a semicolon) define a potential protein-protein interface. MAPPIS will compare only the interfaces defined by the selected chains. Files that are uploaded by the user will be handled in a similar manner.
Note: Single chain protein molecules that have no protein binding partner in the submitted PDB file can not be compared by the MAPPIS method
Stage 2 - Process of MAPPIS:
This window shows the process of activation of MAPPIS. The
five main stages are presented and those that are complete are checked
in the checkbox.
In most of the cases the most time consuming stages are the
construction of the surfaces.

Note: The user does not have to wait for the process to finish, since he/she will receive an email with the link to the output.
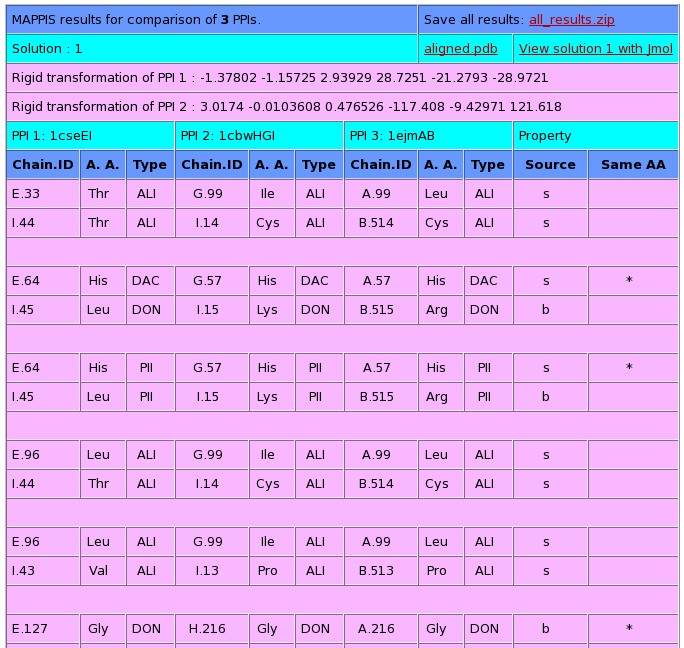
Stage 3 - Output of MAPPIS:
The window below presents the output of the MAPPIS algorithm.
Each pair of rows details the interacting pseudocenters
(physico-chemical properties) of two PPI chains. Each three columns
present the details of a specific PPI: (i) chain identifier and
residue number; (ii) residue type; (iii) pseudocenter type, which can
be donor (DON), acceptor (ACC), mixed donor/acceptor (DAC),
hydrophobic aliphatic (ALI) or aromatic (PI). The next column presents
the origin of the feature: backbone(b) or side-chain(s) if it is the
same for all the matched pseudocenters. The last column details
whether all the pseudocenters were initiated by amino acids with the
same identity.
The 5 top ranking solutions are presented. Each solution can be
viewed with Jmol by clicking on the "View solution with Jmol". In
addition the PDB file with the superimposition of all the complexes
can be downloaded as a separated file (aligned.pdb).
The details of the following column fields can be found below
(click to jump to a description):
Chain.ID
AminoAcid
Property
Source
Dist
Conserved AA
Chain.ID:
The protein chain, followed by the
identity of the amino acid
AminoAcid:
The one
letter amino acid code. However it must be noted that the method is
based on the physico-chemical properties and does not consider the
identity of the amino acids. These are only displayed for the
convenience of analysis.
Property:
The
physico-chemical property that is matched by the algorithm. The method
is based on a representation of each amino acid of a protein as a set
of features that are important for its interaction with other
molecules. The abbreviations of these features are:
DON - Hydrogen bond donor
ACC - Hydrogen bond acceptor
DAC - Hydrogen bond donor and
acceptor (e.g in histidine)
ALI - Aliphatic Hydrophobic property
PII - Aromatic property (pi
contacts)
Source:
This field specifies whether the matched
property is contributed by the backbone or the side-chain of the amino
acid.
The abbreviations are:
b - feature contributed by the backbone
s - feature contributed by the
backbone
Dist:
The distance in space measured
between the matched features.
Conserved
AA:
Marks the features shared by the two molecules that are
contributed by residues with the same identity of the amino acid.
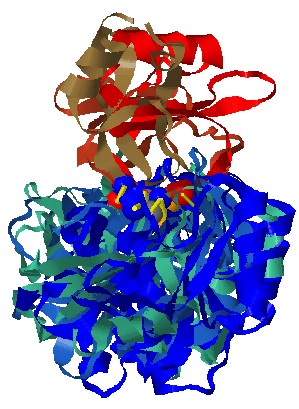
Stage 4 - Jmol options:
Each solution
can be visualized with Jmol. The default view presents the
superimposition of all the complexes as well as the matched
interactions and their physico-chemical properties. The molecules are
colored by model (according to the order of input molecules, the first
is red and blue, the second is lighter red and lighter blue etc.). The
physico-chemical properties (pseudocenters) are represented as balls
and the common interactions are represented by yellow bars. The
matched properties are represented only for the first PPI. Hydrogen
bond donors are blue, acceptors - red, donors/acceptors - green,
hydrophobic aliphatic - orange and aromatic - gray. The interactions
are represented by yellow bars.
surface points that have a similar location in all the
molecules are represented as smaller dots with the same coloring.
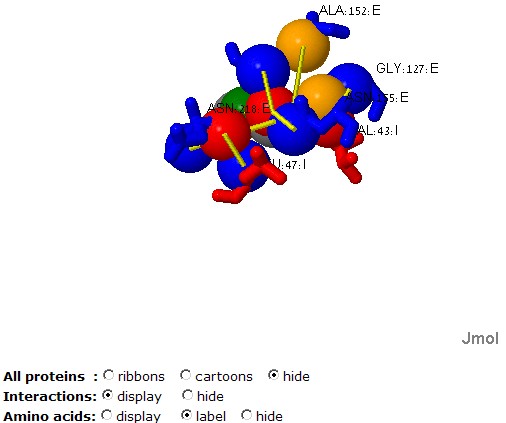
The "Radio groups" at the bottom allow to hide the protein molecules
and focus on the physico-chemical properties, the ligands or the amino
acids.
Please don't hesitate to contact if you have further problems or questions: shulmana@tau.ac.il
Reference: Shulman-Peleg A, Shatsky M, Nussinov R. and Wolfson H.J. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007 Oct 9;5(1):43