|
Biological
Motivation:
There are examples of
proteins that have totally different overall sequences and folds, but
create similar interactions with their corresponding binding partners,
which may also have different overall sequences and folds.
On of the most well studied examples of such interfaces are those
created by proteins of the functional family of serine proteases,
which belong to two different structural folds, trypsin-like and
subtilisin-like, but perform similar functions and have similar
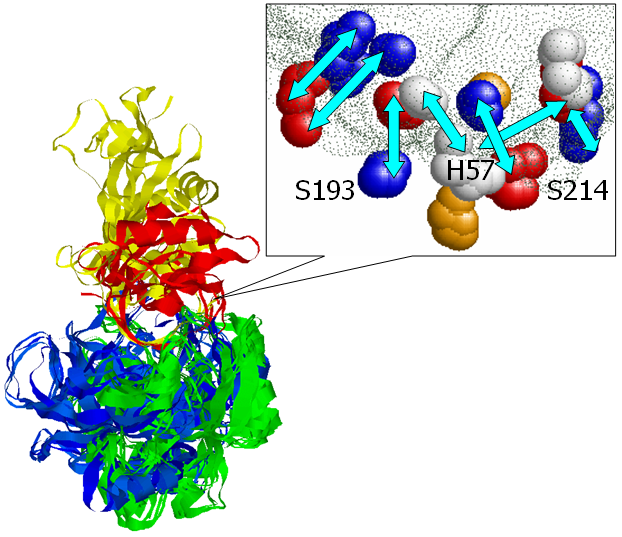
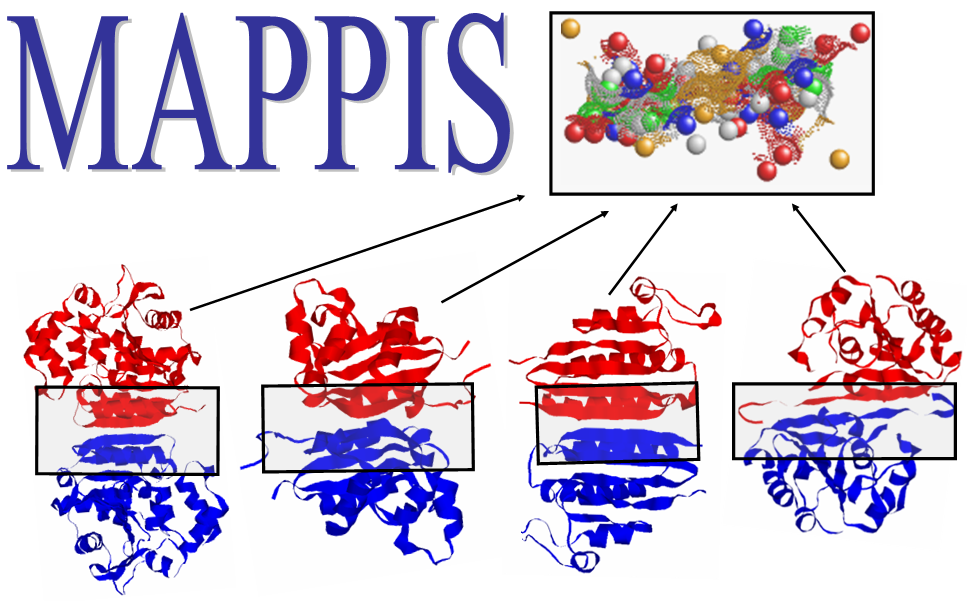
catalytic residues. The figure presents an alignment, obtained by
MAPPIS, between 6 PPIs of serine proteases, with proteins from two
different folds Trypsin-like serine proteases (4sgb, 1ppf, 1acb, blue
and red) and Subtilisin-like (1cse, 2sic, 1oyv, green and yellow). The
rightmost figure presents the common physico-chemical properties and
the conserved interactions between them. The residues that are
conserved in sequence in all the proteins are annotated according to
PDB:4sgb. The MAPPIS solution is correct due to the correct alignment
of the catalytic residues of these proteins. The advantage of MAPPIS
is in the analysis of similarity of the created interactions, which
are presented in the rightmost figure.
|
 |


 Alexandra Shulman-Peleg
Alexandra Shulman-Peleg