
| [Home][Help][About RsiteDB][RsiteDB Classification] |
| Introduction: There are two main types of protein-RNA interactions: (1) Interactions with the backbone of double-stranded RNA molecules and (2) interactions of single-stranded RNA bases that are accommodated in the protein binding pockets. | |
The goal of this database is to describe,classify and predict the interactions between protein binding sites and single-stranded RNA bases. Specifically, RsiteDB describes the protein binding pockets that accommodate extruded nucleotides not involved in RNA base pairing. We define a nucleotide binding site by the protein Connolly solvent-accessible surface area within 2A from the surface of the RNA nucleotide ring (green dots in Figure A). Nucleotides with a protein binding site area larger than 3A are considered as protein interacting. Given a pair of extruded consecutive nucleotides that interact with the protein, a dinucleotide binding site is defined by the pair of the corresponding nucleotide binding sites. The physicochemical properties of the protein binding site amino acids are represented by points in 3-D space termed pseudocenters (see Figure B). Each pseudocenter represents a group of atoms according to the interactions in which it may participate: hydrogen-bond donor, hydrogen-bond acceptor, mixed donor/acceptor, hydrophobic aliphatic and aromatic contacts. We consider only nucleotide and dinucleotide binding sites with more than three surface-exposed pseudocenters. | 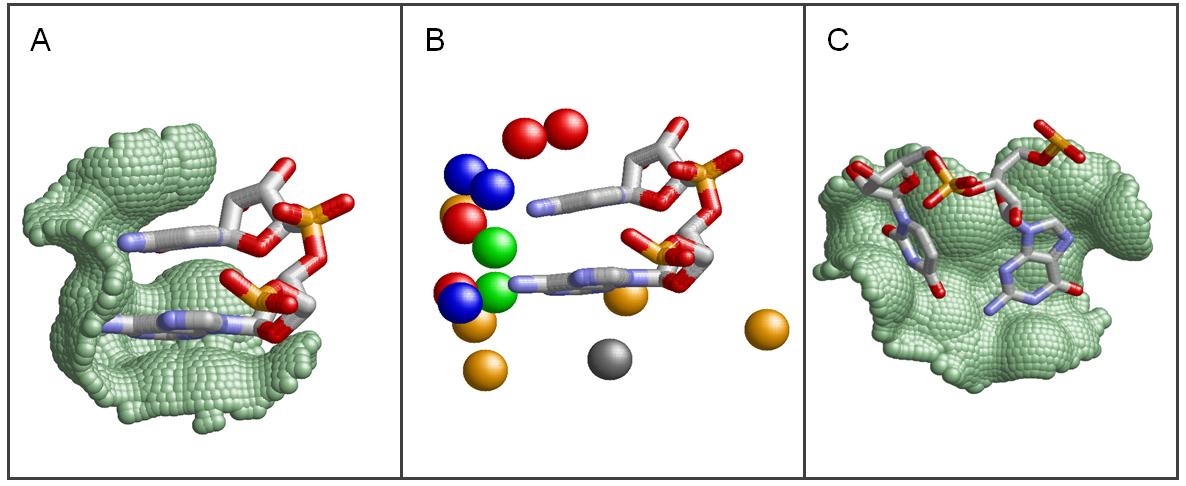 |
| Here we classify all the nucleotide and dinucleotide binding sites and create a non-redundant set of the 3-D binding patterns, which describe their main types of interactions. We consider all the dinucleotide binding sites that accommodate pairs of consecutive extruded nucleotides. In addition, binding sites of single nucleotides that are not a part of such pairs are considered as single-nucleotide binding sites. The classification is based on the novel methodology that validates the cluster quality by multiple spatial binding-site alignment. If the multiple similarity measured by the score of the common physicochemical binding pattern, is lower than a predefined threshold (e.g., 30% of one of the binding sites), the new member is ignored and not added to the cluster. | 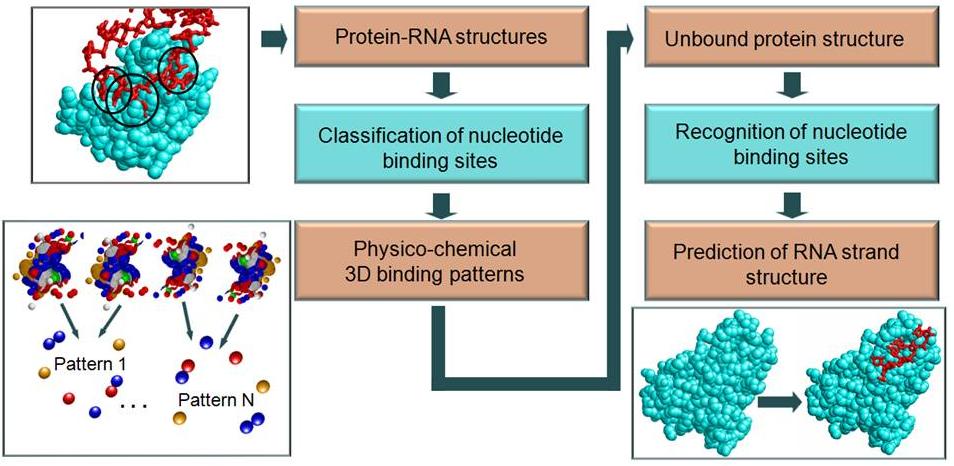 |
| RsiteDB functionality: RsiteDB has two modes of operation. Analysis and classification of protein-RNA interactions: Given a protein-RNA complex RsiteDB analyzes its nucleotide and dinucleotide binding sites. It details the properties of the protein binding pockets that accommodate these extruded nucleotides and presents a list of proteins with similar binding pockets. These proteins may have a totally different overall sequences and structural folds. RsiteDB details and visualizes the features shared by all the binding sites classified to the same cluster. Prediction of RNA dinucleotide binding sites: Given a target, potentially unbound, protein structure we search its surface for regions similar to the created 3-D consensus binding patterns of RNA dinucleotides. The recognized regions are predicted to serve as binding sites. Using leave-one-out tests, the success rate of these predictions was estimated to be about 80%. It must be noted that currently we do not aim to predict whether a protein can bind RNA; rather, given an unbound RNA binding protein, our goal is to predict its binding sites and their modes of interaction. In addition, due to a low number of single nucleotide clusters, currently, we do not use them for the prediction. | |